It has been over 30 years since DNA methylation was first postulated to be a heritable modification capable of influencing gene expression35,70. The addition of a methyl-group to the cytosine base does not change the primary DNA sequence and is therefore considered to be an epigenetic modification, literally meaning to act “on top of” or “in addition” to genetics. The Human Epigenome Project was initiated as a mixed academic and industrial consortium in Europe, aiming to “identify, catalogue and interpret genome-wide DNA methylation patterns of all human genes in all major tissues.” While DNA methylation is commonly agreed to be an epigenetic mark, other modifications of the chromatin structure remain more controversial44. Part of this discussion may have been sparked by the recent support for epigenetics from the NIH Roadmap Initiative. The NIH project definition includes: “Epigenetics is an emerging frontier of science that involves the study of changes in the regulation of gene activity and expression that are not dependent on gene sequence. For purposes of this program, epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable.” Looking at the controversy and concerns raised50it becomes rapidly clear that arguments over the relevance of such a project are largely semantic34,44.
Note: If you’d like to print out a hard copy of these reviews, you can find it at Zymo Research’s website in their Publications.
Epigenetic Mechanisms in Mammalian Genomes
In principle, any mechanism that provides regulatory information to a genome without altering its primary nucleotide sequence could be considered epi- (on top of) genetic. We begin by providing a general overview of the modifications and responsible enzymes, followed by a more detailed discussion of various epigenetic mechanisms that exist in complex genomes and some concluding remarks about the future of the human epigenome project.
DNA Methylation in Development
In mammalian genomes DNA methylation exists primarily within the context of the CpG dinucleotide. Three catalytically active enzymes, DNA methyltransferase 1 (Dnmt1), Dnmt3a and Dnmt3b are required for the establishment and maintenance of DNA methylation patterns8. Two additional enzymes, Dnmt2 and Dnmt3L, display high homology and are expressed in several cell types including embryonic stem cells7. However, deletion of the Dnmt2 gene has no apparent effect62 and its role as a DNA methyltransferase has been questioned28. The Dnmt3L protein lacks the catalytic domain, but is highly expressed in the early embryo, ES cells and germ cells. Dnmt3L deficient mice are viable, but male mice are sterile and heterozygous offspring of homozygous females die due to imprinting defects9,10. In the presence of the de novo methyltransferases, Dnmt3a and Dnmt3b,low levels of non-CpG methylation have been reported20,30,54,69. However, in contrast to plants, the role of non-CpG methylation has not been extensively studied in higher organisms, though it has been suggested that CpA methylation is involved in the regulation of enhancers that are required for olfactory receptor choice in the mouse brain48. Features of DNA methylation in plants, including CG, CHG and CHH (H being A, T or C) methylation, are discussed in the review “Epigenetic Regulation of Gene Expression in Insects and Plants,” Genomic DNA is packaged in higher structures (see page 8, Figure 2).
Histone Modifications in Development
Within a nucleosome 147 bp of DNA are wrapped around a histone octamer (H3, H4, H2A and H2B), the terminal tails of which are subject to many modifications, including methylation, acetylation, phosphorylation, and ubiquitination5,51. Some of these marks (H3K27me3 and H3K9me3) clearly demonstrate mitotic inheritance14,31 while others, including H3K36me3 and acetylation marks, have known regulatory functions, but have unknown heritability5,56. Many of the enzymes regulating such chromatin modifications are known and have been extensively studied including histone acetyltransferases, deacetylases, methyltransferases and more recently histone demethylases75. Two important and well-conserved families, the Trithorax and Polycomb, have SET domain containing histone methyltransferases specific for H3K4me3 and H3K27me3, respectively. The polycomb repressive complex 2 (PRC2) consists of three subunits Suz12, Ezh2 and Eed. The function of Eed and Suz12 remain unknown, whereas Ezh2, is the histone methyltransferase that contains the catalytic SET domain5. PRC2 establishes and binds H3K27 methylation patterns, which are believed to serve as a “docking site” for PRC1 (Ring1, Bmi1, Mel-18 and Cbx family proteins)5.
The Role of Epigenetic Marks During Development
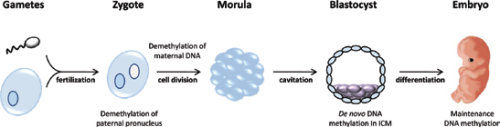
DNA methylation is generally considered a repressive modification and associated with gene silencing8. It is essential for normal mammalian development and required in all somatic cells (Figure 1). In mice, loss of the maintenance methyltransferase Dnmt1 results in embryonic lethality around day E8.5-946 and Dnmt1 mutant embryos display only ~1/3 of the normal levels of DNA methylation. Dnmt1 deficient embryos show rudiments of the major organs, but they are smaller and the embryo itself appears developmentally delayed46. Dnmt3b mutant embryos appear to develop normally before E9.5, but show multiple developmental defects later and no viable term embryos are recovered61. In contrast, Dnmt3a deficient mice can develop until term, but become runted and die about one month postnatally61. Using antibodies against methylcytosine as well as several locus specific DNA methylation assays, it has been established that the paternal genome is actively demethylated prior to the first cleavage division41,52,64,73. The maternal genome, presumably through a passive mechanism, is subsequently demethylated73. Several exceptions to this simplified model exist, including imprinted genes and repetitive elements43.
The paternal genome is demethylated prior to the first cell division of the zygote; maternally derived DNA is demethylated after several cleavage divisions. De novo methylation occurs in the inner cell mass (ICM) cells, which later differentiate into the embryo. Maintenance DNA methylation retains methylation patterns as differentiated cells undergo mitosis.
DNA methylation levels are low in the preimplantation embryo73 and are unlikely to change notably in the absence of Dnmt1. It is therefore not clear why the Dnmt1 mutant embryos are able to initiate development and commit to organogenesis before around day E8.5.Conditional deletion of Dnmt3a results in imprinting defects in the germline and mimics the phenotype of Dnmt3L mutants, suggesting an involvement of both enzymes in the process42. The direct interaction of both Dnmt3a and Dnmt3L has recently been demonstrated63. In this model, Dnmt3L guides de novo methylation to genomic sites that lack H3K4 (me1, me2 and me3) methylation. However, it is unclear whether this mechanism is applicable outside of germ cells. Loss of any one of the PRC2 subunits results in severe gastrulation defects, suggesting an essential role in normal development26,60,67. Similar to PRC2 mutant mice, loss of PRC1 components, such as Ring1b (Rnf2), causes an early embryonic lethal phenotype32. Bmi-1 null mice display several hematopoietic and neurological abnormalities81. Taken together, these data clearly establish the essential role that DNA methylation and Polycomb mediated epigenetic marks play in normal development.
Epigenetic Marks in Differentiated Cells
It has been shown, using conditional alleles, that loss of Dnmt3a in mouse embryonic fibroblasts (MEFs) has no effect, while loss of Dnmt3b results in premature senescence or chromosomal instability as well as spontaneous transformation20. Conditional deletion of Dnmt1 in MEFs causes global loss of DNA methylation and p53-dependent apoptosis38. The global hypomethylation leads to widespread gene expression changes (>600 genes were altered using an array covering 10% of all mouse genes) including imprinted genes and germline specific genes38, the latter of which tends to remain active and unmethylated in ES cells, but methylated in somatic cells55,82. Furthermore, conditional depletion of Dnmt3a and Dnmt3b in hematopoietic stem cells (HSCs; CD34- LSK-) suggested a role of de novo methylation in HSC self-renewal, but not differentiation79. Dnmt1 has been conditionally depleted in the developing nervous system and postmitotic neurons. While the postmitotic neurons did not show DNA hypomethylation or any notable phenotype, the CNS deletion resulted in postnatal lethality of high contribution chimeras or in selective loss of mutant cells in viable low contribution chimeras25. Finally, Bmi-1 has been shown to be required for maintenance of HSC and neural stem cell (NSC) self-renewal57,65,66.
Epigenetic Marks in Pluripotent Cells
Embryonic stem (ES) cells or more generally pluripotent cells represent a developmental ground state that is maintained by a complex autoregulatory network of transcription factors that include Oct4, Sox2, and Nanog40,76. This ground state is of great interest because pluripotent cells have a very unique epigenetic signature and are largely unaffected by the loss of DNA methylation and certain histone modifications16,37,46,54,61,80. Although DNA methylation deficient ES cells cannot readily differentiate, they do maintain pluripotency and contribute to germline competent chimeras upon restoration of Dnmts36,37. PRC2 deficient ES cells show global loss of H3K27me3 and are more prone to differentiation11,16,26,60,67. However, while it had been previously suggested that PRC2 is essential to repress developmental genes and execute differentiation programs11,26,60,67, recent work from Magnuson and colleagues suggests that PRC2 is not required to maintain pluripotency16. Low and high passage Eed -/- ES cells generated early embryonic chimeras with contribution to all germlayers, However, no Eed -/- MEFs could be derived and contribution was rare in late gestation (beyond E12.5 dpc) embryos. Since Eed is required for proper PRC2 assembly, this suggests an essential role in differentiation, but not pluripotency. While the loss of epigenetic marks does not diminish pluripotency, it does nonetheless play a role in regulating gene expression within pluripotent cells. The transcription factor Elf5 plays a central role in the regulation of trophectoderm development and is highly methylated and repressed in undifferentiated ES cells59. Murine ES cells cannot normally differentiate into trophectoderm, however, Dnmt1 mutant ES cells that have lost the ability to maintain Elf5 methylation gain the ability to differentiate effectively into trophectoderm59. Using murine ES cells deficient for all three methyltransferases54 Fouse et al. demonstrated upregulation of over 350 genes in the absence of DNA methylation in the mutant ES cells. The authors found minimal overlap between these upregulated genes and polycomb and/or Oct4, Nanog target genes, suggesting an independent class of genes regulated by methylation27. During initial ES cell differentiation, Oct4 and Nanog need to be repressed. Although DNA methylation is not essential for silencing Oct4, it is required for stably maintaining the repressed state24.
Epigenetic Regulation of Complex Mammalian Genomes
Promoters
Mammalian RNA polymerase II promoters can be separated into low (LCP), intermediate (ICP), and high CpG promoters (HCP)56,74,82. Nearly all HCPs are enriched for H3K4me3 in ES cells and devoid of DNA methylation55,56. Around 22% of these also enrich for H3K27me3 and exhibit a so-called bivalent chromatin structure. Bivalent domains are enriched for key developmental transcription factors and tend to be either not or poorly expressed6. Many of these resolve into a univalent state upon differentiation, though some remain bivalent in certain cell types. These remaining bivalent domains correlate well with gene expression profiles from the adult tissues where they should ultimately become activated56. LCPs rarely contain H3K4me3 (only 6.5%) and frequently show high levels of DNA methylation55,82. Using high throughput bisulfite sequencing, we compared the DNA methylation patterns of ~1 million CpGs across 17 different samples55. Several hundred HCPs had lost H3K4me3 and gained DNA methylation upon differentiation. Importantly, these changes were solely due to the extended culture and not recapitulated in vivo55. In fact, most of the DNA methylation changes associated with differentiation were observed at distal putative regulatory regions 1 to >100 kb away from known promoters.
Distal Regulatory Regions
Gene expression in large genomes is dependent upon long-range control of gene transcription83. Distal regulatory regions, including enhancers, silencers and boundary elements, are subject to dynamic epigenetic changes during cellular differentiation55. The comparison of murine ES cells and ES-derived neural progenitor cells revealed nearly 20,000 distal sites that lost the active marks H3K4me1 and me2 upon differentiation and a similar number that gained H3K4 methylation. In all cases, the change in histone methylation was accompanied by changes in DNA methylation55. Although the majority of these distal sites overlap with highly conserved regions, their functional relevance needs to be further investigated.
Imprinting
McGrath, Solter, Surani and colleagues showed in the early 1980s that the maternal and paternal genomes are not equivalent and contained allele specific imprinting marks3,53,78. Insulin Growth Factor 2 (Igf2) and its receptor (Igf2r) were the first imprinted genes to be discovered77. A hallmark of imprinted genes is the monoallelic expression and parent of origin specific DNA methylation patterns39. DNA methylation has been shown to be required to maintain monoallelic expression15. Using conditional and reversible deletion of Dnmt1, Jaenisch and colleagues have generated “imprint-free” ES cells and mice36. The fact that most imprints are lost upon transient removal of Dnmt1 suggest that alternative mechanisms are either unstable or insufficient to propagate monoallelic expression36; while chimeric mice from this system are viable, they are prone to develop tumors. Loss of imprinting (LOI) of Igf2 has been shown to increase the frequency of intestinal tumors and is frequently found in the normal mucosa of patients with colorectal cancer72.
Mammalian X-Chromosome Inactivation
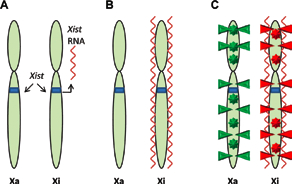
To achieve comparable X-linked gene expression levels in female (XX) cells similar to male (XY) cells (dosage compensation), they have to silence one of their two X-chromosomes1 (Figure 2). X-inactivation occurs shortly after implantation or after differentiation of female ES cells. The up-regulation of a long non-coding RNA, Xist, and the subsequent coating of the inactive X-chromosome is believed to be sufficient for the initiation of X-inactivation1. The spreading of Xist leads to chromosome wide transcriptional silencing and late replication of the inactive X chromosome (Xi). The silencing of Xi is further accompanied by histone modifications (H3K9me3 on the inactive and H3K4me3 on the active X) as well as DNA methylation. In a hierarchical model, the order of events would be as follows: Xist coating of paternal or maternal Xi, late replication timing, histone hypoacetylation, gain of DNA methylation1. Interestingly, the active X (Xa) displays more than two times as much allelic DNA methylation than the Xi. Most of this methylation is found within the gene bodies33.
A) The non-coding RNA Xist is transcribed from the X inactivation center of the inactive X chromosome, Xi. B) Xist binds throughout the length of Xi. C) The silenced Xi displays suppressive histone modifications (red triangles) and DNA methylation at intragenic and promoter loci (red stars). The active X chromosome (Xa) displays activating histone modifications (green triangles) and gene body methylation (green stars).
The fact that all of these genes are biallelically methylated prior to differentiation suggests a mechanism that leads to promoter hypomethylation and gene body hypermethylation33. DNA methylation, histone hypoacetylation, and Xist act synergistically to maintain X-inactivation18. While Xist seems largely dispensable for the maintenance of X-inactivation, DNA methylation and histone deacetylation are essential. Loss of DNA methylation leads to measurable reactivation of the Xi18. Additional evidence for the role of DNA methylation in regulation of X-inactivation comes from the human ICF (immunodeficiency, centromeric instability and facial anomaly) syndrome caused by a germline mutation in the DNMT3B gene45. In contrast to somatic cells, female ES cells maintain two active X-chromosomes. Interestingly, its has been shown that murine female ES cells show global DNA hypomethylation, which might be a result of two active X-chromosomes as well as lower level of Dnmt3a expression85.
Imprinted X-Inactivation
X-inactivation is imprinted during early development in placental mammals1. The paternal X-chromosome is preferentially inactivated during the first lineage differentiation that gives rise to the extraembryonic tissues, whereas X-inactivation in the embryo proper is random. Interestingly, in nuclear transfer experiments X-inactivation in extra-embryonic tissues specifically targets the silenced chromosome of the somatic donor, but remains random in the embryo proper23. When female ES cells with two active X chromosomes were used as donors, the X-inactivation was random in all tissues23. This work suggests that the imprinting marks set during gametogenesis are equivalent to those established during somatic X-inactivation.
Non-coding RNAs and the Mammalian Epigenome
Several large non-coding RNAs that regulate epigenetic modifications in cis (XIST and AIR) or in trans (HOTAIR) have been previously identified and studied12,13,71. A recent genome-wide analysis of K4-K36 domains (H3K4me3 marking the promoter and H3K36me3 marking the transcribed region) revealed a large number of conserved non-protein coding transcripts29. Using published ChIP data, Guttman et al. identified 118 non-coding RNA promoters that overlapped with Oct4 and Nanog binding sites in ES cells. This new data suggests a possible role of large non-coding RNAs, in addition to small RNAs, in the regulation of the complex pluripotency network40. Recently, two groups have shown an additional level of connectivity between non-coding RNAs and epigenetic modifications. The imprinted Air transcript is located in the second intron of Igf2r and regulates its expression in the embryo in cis49. In the placenta, it has at least two additional targets Slc22a3 and Slc22a2. Recent work by Nagano et al. suggests that Air accumulates at the Slc22a3 promoter and directly recruits the H3K9 histone methyltransferase G9A58. Loss or truncation of Air results in loss of imprinting and biallelic expression of the target genes. Air mediated repression of Igf2r in the placenta, however, appears independent of G9A58. The second report has shown the involvement of a non-coding RNA (RepA) in initiation and spreading of X-chromosome inactivation. Loss of RepA results in the failure to induce full-length Xist and recruitment of PRC2 to induce H3K27 trimethylation on the inactive X-chromosome84. Together these examples point to a general mechanism whereby RNAs can guide chromatin-modifying complexes to their specific sites of action.
Sequencing the Human Epigenome(s)
After sequencing dozens of genomes, from fly to human, researchers are now tackling the next frontier: The Human Epigenome. What started as several ambitious projects mostly in Europe is now a major focus of the NIH Roadmap. The initial Human Epigenome Project pilot study focused on DNA methylation and reported single nucleotide resolution data for 253 loci at the human MHC68. The scale-up was completed 2 years later and has provided methylation data for 40,000 CpG dinucleotides (~2,500 loci) across many different samples22. Histone modifications can readily be read on a genome-wide scale using chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq)2,56. Currently two technologies are available that allow the final scale-up to achieve genome-wide DNA methylation analysis21,55. Eventually, shotgun bisulfite sequencing will be affordable across many genomes and this approach has already been successfully implemented in plants17,47. Due to the larger size (3,000 Mb vs. 130 Mb) and the uneven distribution of CpG dinucleotides across mammalian genomes, we have developed a reduced representation approach that allows a reproducible sampling of a smaller, but highly enriched genomic fraction covering ~90% of CpG islands, imprinted regions, highly conserved non coding elements (HCNEs), distal regulatory regions, and repetitive regions55. An alternative approach to nucleotide resolution analyis is termed MeDIP-Seq, which is a combination of methylated DNA immunoprecipitation (MeDIP) and next-generation sequencing4,21. Based on this MeDIP-Seq study, it is expected that up to two mammalian-sized methylomes of ~100 bp resolution can be generated from a single run of an Illumina Genome Analyzer II.
What’s Ahead in Epigenetics
Aided by recent advances in high throughput sequencing technologies researchers have already begun to generate extensive maps of histone modifications and DNA methylation across many mammalian cell types. The coordinated effort of the NIH Roadmap will further assist this endeavor. Changes to the epigenetic state play a central role in many diseases. While cancer is one of the best studied examples, many other pathologies, including diabetes, neurological disorders, and aging may have epigenetic contributions. Analysis of disease state epigenomes will be the second phase of the NIH Roadmap and, when compared with normal reference maps, this data should provide a valuable foundation from which to approach the mechanisms of chromatin mediated genome regulation, DNA methylation and other epigenetic events in mammals. Furthermore, it should provide a general framework for understanding aberrant epigenetic regulation in human development and disease.
Epigenetics References
- Avner, P & Heard, E. Nat Rev Genet 2: 59-67 (2001).
- Barski, A et al. Cell 129: 823-837 (2007).
- Barton, SC et al. Nature 311: 374-376 (1984).
- Beck, S & Rakyan, VK. Trends Genet 24: 231-237 (2008).
- Bernstein, BE et al. Cell 128: 669-681 (2007).
- Bernstein, BE et al. Cell 125: 315-326 (2006).
- Bestor, TH. Hum Mol Genet 9: 2395-2402 (2000).
- Bird, A. Genes Dev 16: 6-21 (2002).
- Bourc’his, D & Bestor, TH. Nature 431: 96-99 (2004).
- Bourc’his, D et al. Science 294: 2536-2539 (2001).
- Boyer, LA et al. Nature 441: 349-353 (2006).
- Brannan, CI et al. Mol Cell Biol 10: 28-36 (1990).
- Brown, CJ et al. Nature 349: 38-44 (1991).
- Cam, HP et al. Nature 451: 431-436 (2008).
- Caspary, T et al. Mol Cell Biol 18: 3466-3474 (1998).
- Chamberlain, SJ et al. Stem Cells 26: 1496-1505 (2008).
- Cokus, SJ et al. Nature 452: 215-219 (2008).
- Csankovszki, G et al. J Cell Biol 153: 773-784 (2001).
- Dodge, JE et al. J Biol Chem 280: 17986-17991 (2005).
- Dodge, JE et al.Gene 289: 41-48 (2002).
- Down, TA et al. Nat Biotechnol 26: 779-785 (2008).
- Eckhardt, F et al. Nat Genet 38: 1378-1385 (2006).
- Eggan, K, et al. Science 290:1578-1581 (2000).
- Epsztejn-Litman, S et al. Nat Struct Mol Biol 15: 1176-1183 (2008).
- Fan, G et al. J Neurosci 21: 788-797 (2001).
- Faust, C et al. Development 125: 4495-4506 (1998).
- Fouse, S et al. Cell Stem Cell 2: 1-10 (2008).
- Goll, MG et al. Science 311: 395-398 (2006).
- Guttman, M et al. Nature (in press).
- Haines, TR et al. Dev Biol 240: 585-598 (2001).
- Hansen, KH et al. Nat Cell Biol 10: 1291-1300 (2008).
- Hanson, RD et al. Proc Natl Acad Sci USA 96: 14372-14377 (1999).
- Hellman, A & Chess, A. Science 315: 1141-1143 (2007).
- Henikoff, S et al. Science 322: 853 (2008).
- Holliday, R & Pugh, JE. Science 187: 226-232 (1975).
- Holm, TM et al. Cancer Cell 8: 275-285 (2005).
- Jackson, M et al. Mol Cell Biol 24: 8862-8871 (2004).
- Jackson-Grusby, L et al. Nat Genet 27: 31-39 (2001).
- Jaenisch, R & Bird, A. Nat Genet 33 Suppl: 245-254 (2003).
- Jaenisch, R & Young, R. Cell 132: 567-582 (2008).
- Kafri, T et al. Genes Dev 6: 705-714 (1992).
- Kaneda, M et al. Nature 429: 900-903 (2004).
- Lane, N et al. Genesis 35: 88-93 (2003).
- Ledford, H. Nature 455: 1023-1028 (2008).
- Li, E. Nat Rev Genet 3: 662-673 (2002).
- Li, E et al. Cell 69: 915-926 (1992).
- Lister, R et al. Cell 133: 523-536 (2008).
- Lomvardas, S et al. Cell 126: 403-413 (2006).
- Lyle, R. et al. Nat Genet 25: 19-21 (2000).
- Madhani, HD et al. Science 322: 43-44 (2008).
- Margueron, R et al. Curr Opin Genet Dev 15: 163-176 (2005).
- Mayer, W et al. Nature 403: 501-502 (2000).
- McGrath, J & Solter, D. Cell 37: 179-183 (1984).
- Meissner, A et al. Nucleic Acids Res 33: 5868-5877 (2005).
- Meissner, A et al. Nature 454: 766-770 (2008).
- Mikkelsen, TS et al. Nature 448: 553-560 (2007).
- Molofsky, AV et al. Nature 425: 962-967 (2003).
- Nagano, T et al. Science (in press) doi 10.1126/science.1163802.
- Ng, RK et al. Nat Cell Biol 10, 1280-1290 (2008).
- O’Carroll, D et al. Mol Cell Biol 21: 4330-4336 (2001).
- Okano, M et al. Cell 99: 247-257 (1999).
- Okano, M et al. Nucleic Acids Res 26: 2536-2540 (1998).
- Ooi, SK et al. Nature 448: 714-717 (2007).
- Oswald, J et al. Curr Biol 10: 475-478 (2000).
- Park, IK et al. J Clin Invest 113: 175-179 (2004).
- Park, IK et al. Nature 423: 302-305 (2003).
- Pasini, D et al. EMBO J 23: 4061-4071 (2004).
- Rakyan, VK et al. PLoS Biol 2: e405 (2004).
- Ramsahoye, BH et al. Proc Natl Acad Sci USA 97: 5237-5242 (2000).
- Riggs, A.D. Cytogenet Cell Genet 14: 9-25 (1975).
- Rinn, JL et al. Cell 129: 1311-1323 (2007).
- Sakatani, T et al. Science 307: 1976-1978 (2005).
- Santos, F et al. Dev Biol 241: 172-182 (2002).
- Saxonov, S et al. Proc Natl Acad Sci USA 103: 1412-1417 (2006).
- Shi, Y. Nat Rev Genet 8: 829-833 (2007).
- Silva, J & Smith, A. Cell 132: 532-536 (2008).
- Stoger, R et al. Cell 73: 61-71 (1993).
- Surani, MA et al. Nature 308: 548-550 (1984).
- Tadokoro, Y et al. J Exp Med 204: 715-722 (2007).
- Tsumura, A et al. Genes Cells 11: 805-814 (2006).
- van der Lugt, NM et al. Genes Dev 8: 757-769 (1994).
- Weber, M et al. Nat Genet 39: 457-466 (2007).
- West, AG & Fraser, P. Hum Mol Genet 14 Spec No 1: R101-111 (2005).
- Zhao, J et al. Science 322: 750-756 (2008).
- Zvetkova, I et al. Nat Genet 37: 1274-1279 (2005).