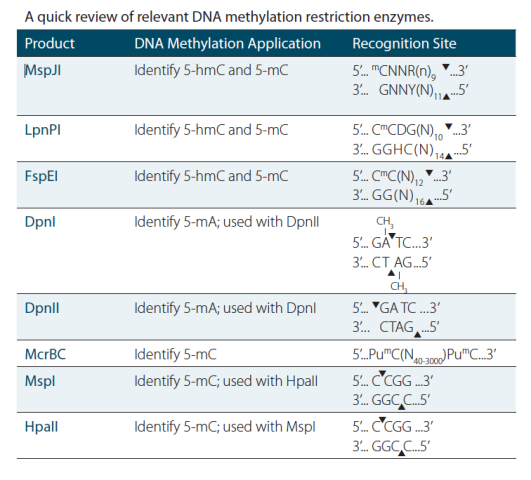
Enzymes and, in particular, restriction enzymes have been invaluable assistants in the laboratory for decades, so it is not surprising that they have been instrumental in driving discoveries in epigenetics-related research as well. Enzymatic manipulation of methylated DNA has seen a tremendous amount of innovation over the last 30 years, making these DNA modifying enzymes great tools to have in your freezer for both today’s experiments and tomorrow’s innovation.
Download Print-Friendly Version >>>
MspI and HpaII
Long before bisulfite conversion, and pre-dating the advent of PCR, restriction enzymes were used to differentiate between DNA samples for their DNA methylation status. MspI and HpaII were an early duo that was used in some of the first DNA methylation analysis methods, which relied upon radioactive labeling for detection (1) and restriction landmark genome scanning, or RLGS. (2)
This pair of restriction enzymes has been particularly powerful as they recognize the same DNA sequence, but exhibit different sensitivities to DNA methylation modifications. This enabled locus-specific discrimination between methylated and unmethylated DNA sequence, ushering in a new era of epigenetics.
Today, MspI and HpaII are still going strong in protocols like the HELP assay (3), which made its debut in 2006 and, more recently, Methyl-Seq (4), which uses the restriction enzyme duo to pre-treat DNA samples before high-throughput sequencing. Both of these approaches provide broad coverage of DNA methylation marks with excellent levels of sensitivity.
Another simplified protocol that has been successful using smaller amounts of input genomic DNA samples, from both FFPE and other clinical tissue sources, is Reduced Representation Bisulfite Sequencing, or RRBS (5).
This protocol uses MspI in an upstream digestion to decipher genomewide DNA methylation studies.
Most recently, MspI and HpaII were incorporated into the EpiMark® 5-hmC and 5-mC Analysis Kit from New England Biolabs (NEB), a novel system that enables the simultaneous detection of both 5-mC and 5-hmC marks (6). The simple protocol leverages the different digestion properties of MspI and HpaII, combined with PCR, to quantitate the amount 5-mC and 5-hmC in a diverse range of samples, using minute amounts of input DNA.
 An overview of the MspI- and HpaII-powered protocol in the EpiMark® 5-hmC and 5-mC Analysis Kit.
An overview of the MspI- and HpaII-powered protocol in the EpiMark® 5-hmC and 5-mC Analysis Kit.
McrBC
A more recently discovered enzymatic method uses McrBC, in which the enzyme cleaves between two methylated cytosines, either GmC or AmC. McrBC, isolated from E. coli, can cleave these sites, even when close together (up to 55 bp apart), which makes it an ideal enzyme for investigating locations that are densely methylated.
McrBC digestion was combined with microarray analysis back in 2005, by Martienssen and colleagues (7). Using McrBC, this team removed the methylated fraction of the genome, and then co-hybridized it with an undigested sample on high-density microarrays, enabling them to identify regions of differential DNA methylation status.
In 2009, Irizarry and colleagues (8) incorporated McrBC into a new approach called Comprehensive High-throughput Arrays for Relative Methylation (CHARM). This approach uses high-density arrays as well, but also uses a novel “smoothing algorithm” to more accurately measure methylation from raw microarray data.
MspJI
Scientists at New England Biolabs recently identified MspJI, one of the newest DNA methylation dependent restriction enzymes for DNA methylation research studies. MspJI, covers a family of enzymes that are related to the known Mrr restriction enzymes in certain bacteria. The MspJI family, first described in 2011, cleaves methylated cytosine when it is two nucleotides away from adenine or guanine, and leaves a four-base overhang on the 5’ side (9). This enzyme was recently used to sequence and map DNA methylation patterns in lung fibroblast genomic DNA, highlighting its potential in genome-wide applications (10).
Additional DNA Methylation Restriction Enzymes Resources
In this guide, we highlight just a few of the key enzymes that have been instrumental in driving discoveries in DNA methylation studies. For more information, check out these useful resources:
- REBASE: a collection of information about restriction enzymes, methylases, the microorganisms from which they have been isolated, recognition sequences, cleavage sites, and methylation specificity.
- NEB Online Resources: a collection of online databases, interactive web tools, and extensive information about restriction enzymes, including their application in epigenetics research, can be found on our website.
DpnI and DpnII
DpnI and DpnII are classic, type IIM restriction enzymes. They were discovered in Diplococcus pneumoniae in 1975, when they were considered unique because they cleaved DNA only at a methylated DNA sequence (11). DpnI and DpnII recognize the same sequence, but different methylation patterns and can be useful in instances where researchers would like to discern two sample populations based on their DNA methylation status. DpnI cleaves methylated adenine to thymine in the 5’ to 3’ direction, while DpnII cleaves the same location, but only when adenine is unmethylated.
Validated DNA Methylation Restriction Enzymes for Today and Tomorrow
These methylation-dependent restriction enzymes, which have been tested and, where relevant, optimized for epigenetics research applications (EpiMark-validated, in the case of NEB’s products ) expand the potential for mapping epigenetic modifications and simplify the study of both 5-mC and 5-hmC marks.
These enzymes have been used to drive innovation in protocols over the years, but they are great tools to consider for further developing protocols that address the future experimental challenges in DNA methylation analysis. To learn more about these enzymes, or to discuss any technical specifications, NEB’s technical services group is always available for support.
1. Cedar, H., et al. (1979) Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 6(6): 2125–2132.
2. Takamiya T., et al. (2006) Restriction landmark genome scanning method using isoschizomers (MspI/HpaII) for DNA methylation analysis. Electrophoresis. Jul;27(14):2846-56
3. Oda M., et al. (2009) High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res.Jul;37(12):3829-39.
4. Brunner AL., et al (2009). Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. Jun;19(6):1044-56.
5. Gu H., and Bock C., et al (2010) Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. February; 7(2): 133–136.
6. Diep, D. and Zhang, K. (2011) Genome-wide mapping of the sixth base. Genome Biol. 12, 116.
7. Martienssen R., et al. (2005) Profiling DNA methylation patterns using genomic tiling microarray. Nature Methods 2:219-224.
8. Irizarry, R., et al. (2008). Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Research, 18: 780-790.
9. Zheng Y., et al (2010) A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. Sep;38(16):5527-34
10. Cohen-Karni, D., et al. (2011). The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. PNAS 108(27)
11. Lacks S., et al. (1975) A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. Jun 10;250(11):4060-66