Whether you’re the head of a lab, household, or leading a country, budgets are a reality. Like those old jeans you can’t seem to donate, they seem to get tighter every year. So when slick, new methods surface that tackle some tough challenges like single nucleotide resolution 5-hmC profiling, researchers’ excitement is quickly tempered by the experimental costs required to execute the latest and greatest technique. A nice workaround to this issue can be achieved by combining 5-hmC enrichment sequencing with restriction digestion-based targeted validation.
5-hmC Enrichment Sequencing
The enrichment sequencing process is not much different than your standard ChIP sequencing workflow:
- Fragment sample and ligate adapters
- Enrich 5-hmC DNA from the rest of the 5mC and non-methylated DNA
- Sequence away
- Map peak and perform other bioinformatics wizardry
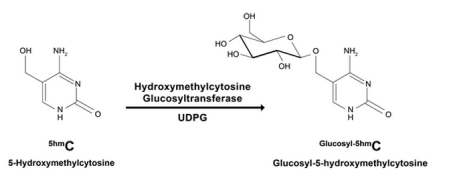
The performance of this approach is largely dependent on the quality of your enrichment. An effective approach that has been working for Zymo Research is to first convert 5-hmC to β-glucosyl-5-hmC (5-ghmC) using T4 β-glucosyltransferase. This sets you up to pull-down all 5-ghmC using J-binding protein 1 (JBP1), which has a strong affinity for 5-ghmC.
Cutting to the Chase with Restriction Digestion
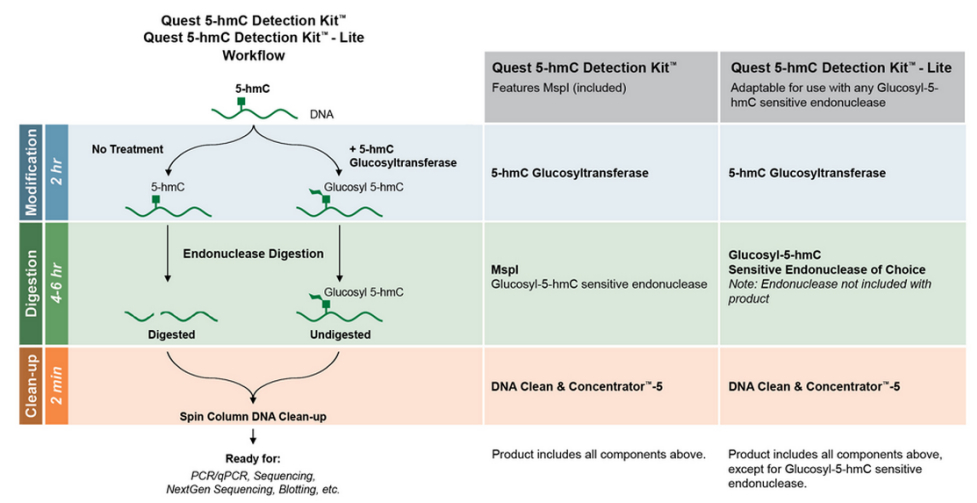
JBP1 sequencing is a great first step that’s specific and pretty user-friendly, but it will only get you down to the regional view of where 5-hmC marks may exist. In many cases this resolution might suffice, but for those looking for more precise mapping of 5-hmC marks, restriction digestions can help.
“After our genome-wide scans, we go in and validate this data using our restriction digestion approach to validate loci of interest. This way we can analyze more of the epigenome at a price that is reasonable for most labs,” stated Marc Van Eden, Vice President of Marketing at Zymo Research.
Another nice aspect of validating with restriction digestion assays is that you’re coming at the validation with a different approach entirely: binding proteins vs. restriction digestion, so when the data line up, you can be that much more confident in your results.
Restriction digestion-based analysis of 5-mC is nothing new. Researchers have been using methyl-sensitive and –dependent enzymes for years, but recently talented researchers have put glucosyl-5-hydroxymethylcytosine sensitive restriction endonucleases (GSREs) to work to obtain more precise mapping of 5-hmC presence.
GSREs are blocked by the presence of 5-ghmC. Since the 5-hmC DNA in JBP1 sequencing has already been converted to 5-ghmC, you’re already at Step 2 in the validation workflow. All that’s left is digestion and clean-up, before heading into your preferred validation approach like qPCR or targeted sequencing.
Breakthroughs without Breaking the Bank
Don’t let the pesky practicality of a budget hold you back from your next breakthrough discovery, combo up 5-hmC enrichment with GSREs and see what surprises you uncover. Not feeling up to that task? Check out Zymo Research’s Genome-wide 5-hmC Analysis Service. You provide the sample, they provide the data.