The nightly news lately makes it seem like the world has gone upside down, and now we’ve got some science to back it. Honglei Zhou and Isidore Rigoutsos from Thomas Jefferson University recently revealed that mammalian miRNA targeting doesn’t always limit itself to 3′ miRNA action. It now seems that in addition to the traditional 3’UTR targeting, mammals are starting to look a bit more like plants as they also target a number of the 5′ untranslated region (5′ UTR) and amino acid coding sequence (CDS) sites.
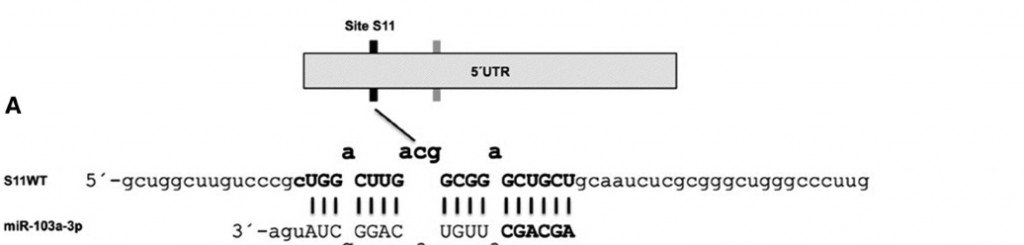
To help the notion of 5′ UTR to catch some steam the team characterized two (miR-103a-3p) target sites in the 5′ UTR of GPRC5A, a two-faced cancer gene. Here’s what they found:
- The interaction of miR-103a-3p with the 5′ UTR targets reduces the expression levels of both GPRC5A mRNA and GPRC5A protein.
- This naturally occurring miRNA target of the 5’UTR goes down in a seed-depedent manner.
- Interestingly, by ectopically expressing miRNA sponges that contain 5′ UTR targets they are able to reduce miR-103a-3p levels and increase GPRC5A mRNA and protein levels.
Overall, this research offers a rare glimpse into the post-transcriptional regulation of a tumor suppressor/oncogene and highlights the complexity of miRNA targeting.
Getting Into 5’UTR and CDS target sites
Senior researcher Isidore Rigoutsos shares that “Back in 2006 we provided evidence that CDS and 5’UTR sites are also abundant in addition to the 3’UTR ones. Then we and many other people showed examples of CDS targets starting in 2008 and by now there are a few dozen papers showing CDS targets. 5’UTR targets seemed harder to come by: to date I know of only six reports that show 5’UTR miRNA targeting (in 2 of the six the targeting leads to up-regulation whereas in the other four it leads to down-regulation). In my opinion what has limited the discovery of CDS and 5’UTR targets is not their relative absence compared to 3’UTR targets but the fact that the vast majority of the miRNA target prediction tools focus on the 3’UTR. Our own rna22 algorithm is an exception to this in that it can work with any region of messenger RNA and in fact with any transcript (coding and non-coding).”
Make sure your research is on target in RNA, July 2014