A couple of years ago, researchers keen to analyze 5-hmC had a couple of options, but those early approaches couldn’t quite satisfy all of their increasing demands. Fast-forward a bit and it’s a very different story. Today there’s a plethora of options, each with its own strengths and weaknesses. As a result, one of the biggest challenges researchers currently face in 5-hmC analysis arises long before experiments are conducted: choosing the right tool for the job.
Download Print-Friendly Version >>>
A Brief History of 5-hmC Analysis
Skirmantas Kriaucionis and Nathaniel Heintz kicked off the 5-hmC analysis wave with a Science publication back in 2009, after they noticed an odd spot on their TLC plates while analyzing some Purkinje DNA samples.
In this guide…
- We introduce various methods for global, locus-specific, and single-nucleotide 5-hmC analysis
- Review the strengths of each of the approaches
- Discuss the technical expertise required for each approach
- Summarize the material costs associated with each approach
They quickly deployed both HPLC and MS approaches to confirm that this odd spot was 5-hmC. These TLC, HPLC, and MS-based approaches were accurate methods and they were instrumental in catapulting 5-hmC into the spotlight, but their ability to help researchers get to the bottom of exactly what role 5-hmC played and where it was located was limited.
In this same publication, Heintz and Kriaucionis suggested what later groups would confirm; bisulfite conversion (at least in isolation), and other popular 5-mC analysis methods would not detect 5-hmC at all, or wouldn’t be able to discern it from 5-mC. Opportunistic academic and commercial scientists dove into their toolkits, and within months a variety of new approaches surfaced.
5-hmC Enrichment
The first wave of tools to emerge brought antibodies and other enrichment approaches that could reliably and specifically bind 5-hmC. Two approaches for 5-hmC enrichment that were adopted early-on were:
5-hmC Antibody Enrichment (hMeDIP)
Polyclonal and monoclonal antibodies that could selectively enrich 5-hmC vs. 5-mC were the first on the scene. These provided researchers with some power to discern between the two DNA methylation marks, and were tremendously helpful. Using antibody-based enrichment, Haffner and colleagues (Oncotarget, 2011) showed that 5-hmC was abundant in the majority of adult and embryonic tissues and that 5-hmC levels were markedly lower in many cancer tissues, relative to their normal counterparts. Antibody enrichment sequencing also showed that 5-hmC was prominent in key regulatory regions like enhancers, as well as in gene bodies (Genome Biology, 2011).
J-binding protein 1 (JBP1) 5-hmC Pull-Down
Although antibody-based enrichment worked well for many labs, researchers sought alternative ways to enrich 5-hmC. Most of the subsequent methods took advantage of β-glucosyltransferase (β-GT), a viral enzyme that tags a glucose molecule to the hydroxyl group of 5-hmC to create β-glucosyl-5-hmC (5-gmC).
This led to a number of talented teams publishing ways to enrich 5-hmC without antibodies. Robertson, et al., (Nucleic Acids Research 2011) introduced a clever alternative to antibody-based 5-hmC enrichment. They first converted 5-hmC to β-glucosyl-5-hmC (5-gmC) using T4 β-glucosyltransferase (T4-BGT). They then pulled down the 5-gmC with another protein, J-binding protein 1 (JBP1), which has a strong affinity for 5-gmC. The pulled-down material can be used in quantitative PCR, microarrays, and sequencing.
An optimized version of this protocol exists in Zymo Research’s Quest 5-hmC™ DNA Enrichment Kit, which includes all the reagents for JBP1 pull-down of 5-gmC.
5-hmC-Restriction Digestion
Differential restriction digestion provides researchers with another great way to discern between 5-mC and 5-hmC. Unlike the JBP1 pull-down, this approach introduces glucose to create distinct sample populations that exhibit different sensitivities to restriction enzymes.
Glucosyl-5-hydroxymethylcytosine sensitive restriction endonucleases (GSREs)
New England Biolabs and Zymo Research both introduced restriction-digestion-based systems. These protocols first glucosylate 5-hmC marks, and then use a GSRE (e.g. MspI) that can digest DNA when cytosine, 5-methylcytosine, or 5-hydroxymethylcytosine is within its recognition site in CpG context, but is blocked by the presence of 5-gmC, to create two sample populations.
NEB’s system includes HpaII in addition to MspI. Both enzymes recognize the same sequence (CCGG), but they have different digestion sensitivities. Unlike MspI, HpaII is blocked by all modifications (5-mC, 5-hmC and 5-gmC) in CpG context, so it only cuts where unmodified cytosine is present.
“By using methylation- and hydroxymethylation-sensitive restriction enzymes, researchers can obtain more precise, sequence-specific information when profiling 5-hmC in their target gene. Analyzing the two marks in concert provides real-time, stage-specific data on cellular growth and development, expanding the potential for biomarker discovery.”
-Sriharsa Pradhan, Research Division Head of RNA Biology at New England Biolabs
Combining 5-hmC Enrichment with Restriction-Digestion Methods
A two-step experimental approach for researchers starting out with 5-hmC analysis is to first get a sense of where the interesting 5-hmC hotspots are in the model you’re studying using an enrichment-based analysis, and then move in with a restriction-digestion approach coupled with a qPCR readout.
This takes advantage of the strengths of each method. “For our genome-wide 5-hmC analysis service, we use a JBP-1 pull down to provide a genome-wide look at 5-hmC from minimal input sample. Then we go in and validate this data using our restriction digestion approach to validate loci of interest. This way we can analyze more of the epigenome at a price that is reasonable for most labs,” stated Marc Van Eden, Vice President of Marketing at Zymo Research.
Bisulfite-Based 5-hmC Analysis
From late 2009 until early 2012, scientists continued to develop innovative approaches that worked around the fact that the commonly-used bisulfite-conversion protocols could not discern 5-mC from 5-hmC. The 5-hmC enrichment and digestion-based approaches were very useful, but many researchers still wanted the single-base resolution power that their beloved bisulfite provided.
Two alternatives surfaced in June of 2012: Oxidative Bisulfite Sequencing (oxBS-Seq) and Tet-assisted bisulfite sequencing (TAB-Seq). Both methods use oxidation to create distinct populations within samples, but they accomplish this in different ways. Both also work with high-throughput sequencing platforms to provide quantitative 5-hmC mapping with single-base resolution. At last!
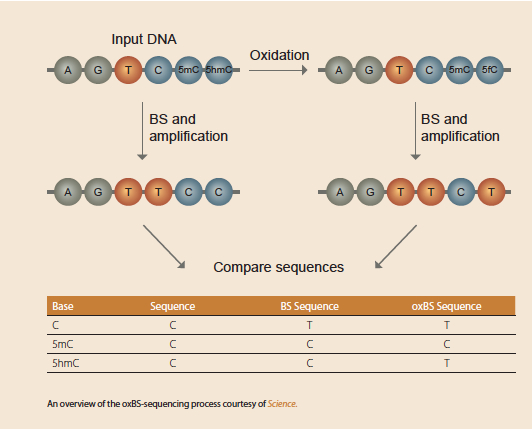
Oxidative Bisulfite Sequencing (oxBS-Seq)
oxBS-Seq, developed by Michael Booth and Miguel Branco, is a smooth protocol that uses a few minor tweaks before performing a standard bisulfite-conversion reaction. The team took advantage of the fact that 5-formylcytosine (the oxidative product and structural relative to 5-hmC) deaminates to uracil under bisulfite treatment, whereas, 5-mC remains unchanged. The team found they could oxidize 5-hmC marks with potassium perruthenate (KRuO4) and convert them to 5fC while leaving unmodified cytosine and 5-mC unchanged.
From here, standard bisulfite conversion converts what was originally 5-hmC (now 5-fC) and unmodified cytosine to uracil and leaves 5-mC untouched. Then, when an oxBS-Seq library is subtracted from a standard BS-Seq run, it’s possible to see the differences between 5-hmC and 5-mC.
In the process of validating the approach, Booth and Branco applied oxidative bisulfite upstream of reduced representation bisulfite sequencing (RRBS) and found that in embryonic stem cells, 5-hmC accumulated in lower-density CpG islands, which usually have intermediate 5-mC levels.
Tet-assisted bisulfite sequencing (TAB-Seq)
TAB-Seq, developed and introduced by Mia Yu, Gary Hon, and Keith Szulwach and their colleagues tackles single-nucleotide 5-hmC mapping from a different angle. The team leveraged previous findings that showed Tet proteins oxidize 5-mC to 5-hmC, but also 5-hmC to 5-caC, and the fact that 5-caC behaves like unmodified cytosines during bisulfite conversion.
In TAB-Seq, 5-hmC marks are first tagged with a glucose using β-glucosyltransferase to create 5-gmC as is done with enrichment methodologies. However, in this case the goal is to protect the 5-hmC marks from further oxidation. After protecting the 5-hmC marks, samples go through enzymatic oxidation with an excess of Tet protein during which all 5-mC is converted to 5-caC.
From here, standard bisulfite conversion converts unmodified cytosines and 5-caC to uracil and 5-caU respectively, but the 5-hmC remains modified as 5-gmC. Sequencing these bases presents the modified 5-hmCs (5-gmC) as cytosine. Comparing TAB-Seq with standard BS-Seq libraries allows precise mapping of 5-hmC vs. 5-mC modifications.
Global 5-hmC Detection
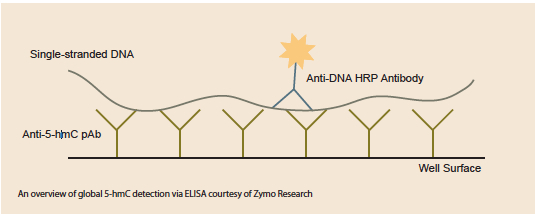
The early 5-hmC studies were made possible through more global detection approaches like HPLC, and MS-based analysis. Since then, there have been many advances that deliver more precise mapping of 5-hmC presence, but there’s also been innovation in global 5-hmC detection as well. Scientists at Zymo Research recently released an ELISA system that they use routinely in their own work. ELISAs are sensitive, simple, and fast. “The ELISA is a great system to quickly assess the levels of 5-hmC in an experimental system. Many groups may use it as a preliminary screen or to identify which systems they want to further study,” added Van Eden.
Terragani et al., also introduced a radioactive labeling assay with β-GT to quantitatively measure 5-hmC levels in a range of tissues.
5-hmC Analysis Techniques Breakdown
So which of these methods will work best for you? That depends. Like most technology landscapes, each method brings different benefits to the bench that may be more or less meaningful depending on skill level, budget, experimental needs, and other factors. Let’s start by comparing these methods in terms of the more common factors that researchers consider when pursuing new methods.
Mapping Resolution
Some researchers need single-nucleotide resolution, while others are happy with a regional view of 5-hmC presence. Here, we’ll take a closer look at how these approaches match up in terms of mapping resolution.
Expertise Required and Technical Barriers to Entry
What good is a method if you can’t get it up and running in your lab? In this section we’ll discuss what it actually takes to get moving with these approaches.
Cost
Funding is tight out there. In this section we’ll outline the rough costs that accompany each of these approaches.
5-hmC Mapping Resolution
Being able to discern between 5-mC and 5-hmC is an essential step forward towards better understanding the role of these marks in development and disease. Many researchers feel that it is essential to map 5-hmC at single nucleotide resolution, while others find that locus-specific detection will suffice. Here’s how these methods compare in mapping resolution.
Single Nucleotide 5-hmC Resolution
When it comes to mapping resolution, the oxidative-bisulfite sequencing methods (oxBS-Seq and TAB-Seq) reign supreme. They are the only methods (as of June 2012) that enable single nucleotide resolution of 5-hmC, so if you no longer need to be convinced of the relevance of 5-hmC and want to take the dive, these are your best bet.
Regional or Locus-Specific 5-hmC Detection
The resolution provided by the 5-hmC enrichment (antibody- and glucosyltransferase-mediated) is dictated by the resulting fragmentation profiles. Although these can vary from a few hundred to a few thousand base pairs, it’s safe to say that they are best used for a more regional assessment of 5-hmC abundance. Restriction digestion approaches will typically provide better resolution than enrichment (antibody or JPB1) though.
Many groups have acquired very insightful data using regional approaches, but if you’re after single-nucleotide resolution, you will want to consider a bisulfite-based approach.
Global 5-hmC Detection
HPLC, LC/MS and ELISA approaches are useful approaches for a quick look at global 5-hmC levels. Due to the nature of these assays, they will not provide much insight into the general location of the 5-hmC marks. In many cases, fine resolution may not be necessary, at least not immediately.
“Some of our customers use global 5-hmC detection approaches first to better understand the general prevalence of 5-hmC in their model system before heading on to more advanced methods. In other scenarios, groups interested in screening compounds for their impact on 5-hmC expression, might opt for a quick global yes/no answer before moving into a more thorough study.”
-Larry Jia, R&D Director at Zymo Research
Technical Expertise Required
You would probably rather spend your time producing results than validating new protocols, so it’s always relevant to discuss the technical barriers and expertise required to bring a new method into your lab.
oxBS-Seq of 5-hmC
The oxidative-sequencing approaches have yet to be “kitted” but both groups say that a commercial version of their method is in the works. At this stage it is difficult to say which of these will be more user friendly. oxBS-Seq uses a chemical, potassium perruthenate (KRuO4), to oxidize, whereas TAB-Seq uses Tet enzyme.
With TAB-Seq, the quality and activity of the Tet enzyme is critical, as it needs to oxidize over 97% of 5-mC to 5-caC. This is not a simple task, but we’re told that a commercial provider will release a highly active Tet for validated this application in the near future. This should remove that barrier to uptake. The rest of the protocol seems fairly straightforward.
With oxBS-Seq, the sample and oxidizing reagent both need to be as pure as possible.
“Our method works well as long as the DNA sample is pure. The chemical oxidation reagent is highly efficient and works in a manner that is insensitive to sequence context. The quality of the oxidant solution makes a difference.”
– Dr. Wolf Reik and Dr. Shankar Balasubramanian, co-developers of oxBS-Seq.
Antibody Enrichment of 5-hmC (hMeDIP)
Pulling down 5-hmC is not much different from other epitopes in that success hinges on the quality of the antibody used and a reproducible hydroxyl-MeDIP (hMeDIP) protocol or kit. If you have a good antibody and kit, it should be smooth(er) sailing; if not, tighten that seatbelt because there will be turbulence ahead. A number of 5-hmC polyclonal and monoclonal antibodies have surfaced in the last couple of years – try to find one that has been validated or qualified in hMeDIP. Many suppliers use different terminology for how they validate an antibody (qualified, validated, ChIP –grade etc.,) Ideally, you will want the specific lot of antibody you’re purchasing to have been validated to work effectively in immunopreciptations, or in an hMeDIP setting.
We asked Michael Sturges of EMD-Millipore to explain a bit more on what to look for in an antibody for hMeDIP: “The amount of 5-hmC DNA found in a given cell is typically lower than that of 5-mC. For really good enrichment of 5-hmC DNA, having an antibody with high affinity and specificity will lead to a better result. We evaluated multiple 5-hmC antibodies to find the best ones. These turned out to be monoclonals. Both show very good specificity and affinity and have been published for hydroxyMeDIP. Because they are monoclonals, we see minimal variation in performance from lot-to-lot, so we’re confident that other labs will be able to replicate the success of the groups who published using these clones.” shared Sturges.
J-binding protein 1 (JBP1) 5-hmC Pull-Down
The original developers put a lot of effort into developing this protocol. They demonstrated via mass spectrometry that β-gt can convert 5-hmC residues to β-glu-5-hmC at nearly 100% efficiency. They also showed that JBP1 showed strong specificity for β-glu-5-hmC. Fortunately, it didn’t take long before a commercial version was released to spare you the hard labor. Zymo Research kitted this protocol into a fairly painless three-step process in their Quest 5-hmC™ DNA Enrichment Kit.
5-hmC-Restriction Digestion
The MspI, and MspI/HpaII restriction digestion approaches available in Zymo’s Quest 5-hmC Detection Kit™ and NEB’s EpiMark® 5-hmC and 5-mC Analysis system respectively, share the same glucosylation step as the JBP1 5-hmC pull-down, but they exchange an enzymatic digestion step for the pull-down. This step doesn’t really create any more or less hands-on time and should be just as easy for first-time users as for more experienced hands. Moreover, the digested target DNA can be easily analyzed with standard qPCR protocols to approximate the amount of 5-hmc present.
Cost of 5-hmC Analysis Approaches
In today’s research economy, how can we ignore the costs associated with using these approaches? In general, the upstream portion of these protocols is pretty reasonable no matter which approach you take. It’s what lies beyond that will make or break your lab wallet. Basically, the cost scales closely with the amount of site-specific 5-hmC information obtained from the method.
5-hmC Sequencing Costs
For the oxidative bisulfite sequencing methods, commercial kits have yet to be released so it’s hard to say what the cost-per-reaction will be. What is easier to say with confidence is that this cost will look negligible next to the sequencing costs. The developers of both of the oxidative-bisulfite sequencing approaches estimate that an entry point of 15 to 50-fold coverage would suffice for higher density 5-hmC regions, but if you want to look at mid- to lower-density 5-hmC regions, brace yourself.
Sure sequencing costs continue to drop but it’s still a huge chunk of the budget, so if you’re looking to head into broad, single-nucleotide resolution 5-hmC sequencing, open up that wallet. But is chasing down quantitative information for every 5-hmC mark even necessary? It’s tough to say what coverage and depth will be necessary at the moment.
“For hydroxymethylation, if you have let’s say one-fifth of the levels of methylation as a genome-average, you will need to sequence more, but how much more also depends on the distribution of hydroxymethylation in the genome. As we get better ideas about the distribution of 5-hmC in each cell type we can make better predictions about the sequencing depth you will need in each case,” added Dr. Reik.
Alternatively, genomic fractionation methods like reduced representation bisulfite sequencing (RRBS) can be applied to reduce the amount of landscape that needs to be sequenced. Dr. Reik’s team combined the oxidative-bisulfite approach upstream of RRBS in their recent Science publication.
Locus-Specific 5-hmC Detection Costs
When you step away from single-nucleotide resolution into regional or locus-specific 5-hmC detection, things get considerably cheaper. Your costs will stem from two areas: the sample preparation approach, which will run $10-20/reaction depending on the vendor and reaction size, plus your standard qPCR reaction costs, which will tack on another $5/reaction. We’re not talking about a major cash outlay unless you are running thousands of samples.
Global 5-hmC Detection Costs
Another reason to consider using a global 5-hmC detection method before moving on to more involved studies is the affordability of global detection. At $2-10/reaction in material costs for running an ELISA, HPLC run, or a radiolabeled assay, it’s probably the cheapest way to gain a quick sense of 5-hmC abundance in your samples, or to monitor variance of 5-hmC in response to a treatment.
Summary
Like any new field, the technology landscape for studying 5-hmC is rapidly evolving. As more questions about the biology of this intriguing mark are answered, the pros and cons of the various methods to study it will change as well. We’ll continue to see more technology innovation and protocol streamlining. Most exciting of all, we’re guaranteed to see fascinating research findings that contribute to a better understanding of 5-hmC in gene expression. Be sure to check out our ongoing coverage of 5-hmC as we add new Headlines and Features highlighting new publications.