Will it rain? Will my team win? Will I get that grant? There are instances where a glance into a fortune-teller’s crystal ball could definitely give you the upper hand. Although the crystal ball is a myth, researchers from the lab of Prabhas V. Moghe (Rutgers University, New Jersey, USA) have recently developed a means to predict the future of single stem cells using a newly developed technique termed “Epi-mark Descriptor Imaging of Cell Transitional States” or EDICTS for short.
This epigenetic necromancy (don’t worry, they didn’t really communicate with the dead!) images global changes to the histone modification patterns (or Epi-marks) which control lineage-specific gene expression programs and uses this information to predict stem cell fate at the single cell level. Specifically, EDICTS employs super-resolution imaging and high content data analytics to visualize patterns of co-occurring trimethylations of lysine residues 4 (H3K4me3) and 27 (H3K27me3) on histone 3 (H3K4K27me3) and quantify nucleosome level changes in single cells. Notably, the combination of these two normally opposing histone modifications, known as a bivalent domain, marks regulatory sequences important to stem cell fate acquisition.
To develop EDICTS, Kim et al. first employed immunoelectron microscopy (IEM) and gold-labelled histone modification-specific antibodies to create visual maps of H3K27me3 (inhibits gene expression) and H3K4me3 (promotes gene expression) in human mesenchymal stem cells (hMSCs) and hMSCs undergoing directed osteogenic and adipogenic differentiation. Although this technique destroyed the cells, IEM combined with image characterization analysis successfully captured phenotype-specific patterns of H3K4K27me3. This creates a data resource which can then be used to assess histone modification patterns acquired with a non-destructive technique and predict single stem cell fate in an experimental sample.
To further “train” the developing system, the authors treated hMSCs with lysine methyltransferases (KMTs) inhibitors to engineer chromatin states with high levels of H3K4me3 or H3K27me3 They then captured histone modification patterns in single cells via a live-cell super-resolution imaging technology termed time-gated stimulated emission depletion (G-STED).
The preparation and training paid off, as the authors successfully captured distinct H3K4K27me3 patterns in cultures of differentiating hMSCs and attributed single live cells to a stem cell, adipogenic, or osteogenic fate. EDICTS also predicted stem cell fate with 100% statistical accuracy (!) in induced pluripotent stem cells differentiating towards different neuronal fates. Importantly, accurate cell fate attribution at the 72-hour mark represents a shorter reporting time than the 2-4 days required for lineage-specific downstream biomarkers such as RNA and protein.
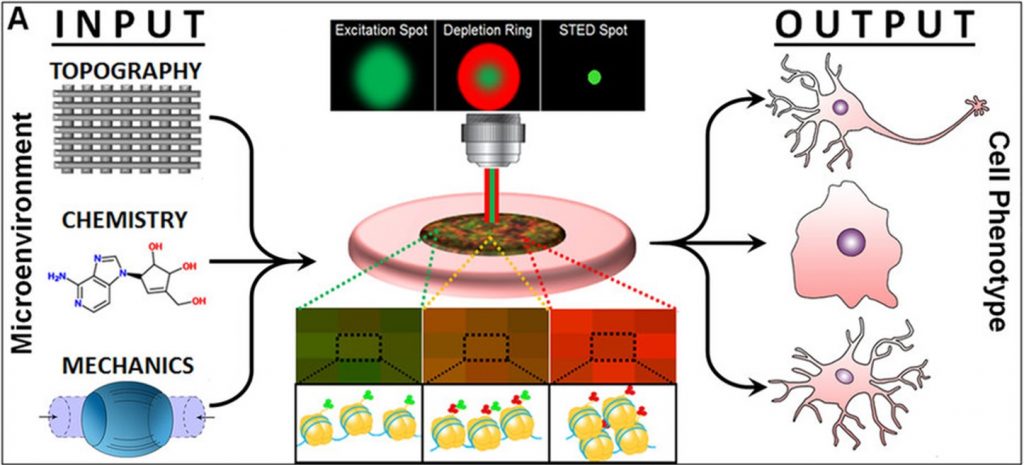
The authors hope that this single cell technique may promote the realization of enhanced differentiation protocols, new lineage-specific inhibitors/growth factors, and biocompatible substrates with different mechanical and topographical properties, all with the goal of constructing better, highly efficient, and cost-effective regenerative therapies (See Figure).
While we don’t have a crystal ball here at Epigenie, we are sure that EDICTS will have a bright future. Read all about it at Sci Rep, Jan 2017.