Move over DNA methylation, take a seat histone modification, it’s time to make room for RNA methylation. When you think of the importance of epigenetics to brain function and cognition, you probably think of well characterized changes DNA methylation, histone marks, and ncRNA. Well you better make some room in your own neural networks, because three new studies have found a role for a crucial new player: m6A.
Methylation of adenosine in RNA to N6-methyladenosine (m6A) has been known for decades; however, its abundance and pathological relevance have only recently been uncovered. N6-methyladenosine (m6A) is the most common internal mRNA modification, present in about one quarter of all RNAs. It increases mRNA stability and affects nearly all mRNA processes. m6A is deposited co-transcriptionally by a methyltransferase protein complex consisting of METTL3, METTL14, WTAP, KIAA1429, and RBM15/RBM15B. m6A is also demethylated by FTO and ALKBH5. RNA methylation acts to dynamically regulate the fate of individual mRNA transcripts. In mammals, m6A appears to control many processes including stem cell proliferation, DNA damage response, X inactivation, and tumorigenesis.
A lot of evidence suggests that like other epigenetic processes, m6A may be particularly important in the brain. m6A is more prevalent in the brain than most other tissues, especially in adulthood. Some interesting work has shown that m6A is dynamic in neurons and responds to experience. m6A levels and patterns differ across brain regions, but all m6A writers and reader proteins are expressed in all brain cell types. Variants in the RNA demethylase genes FTO and ALKBH5 have been associated with several psychiatric disorders. FTO has also been associated with memory formation and regulation of dopaminergic neuronal function.
In this special feature, we summarize three recent studies that have really delved into the role of m6A in the brain using different mouse models. All three used m6A immunoprecipitation (IP) followed by sequencing (m6A-seq) in mouse brain; a challenging approach since very little m6A RNA is present after IP. Each tackled this challenge in different ways.
Image courtesy of Mareen Engel
m6A in Brain Stress Response
The brain’s response to stressful stimuli is a complex molecular cascade involving DNA methylation and histone modification. A better understanding of these mechanisms has applications in understanding acute stress response and the development of stress-related psychiatric disorders. Defining the role of m6A in a specific process such as stress response would facilitate a clearer picture of its functional role in the brain in general.
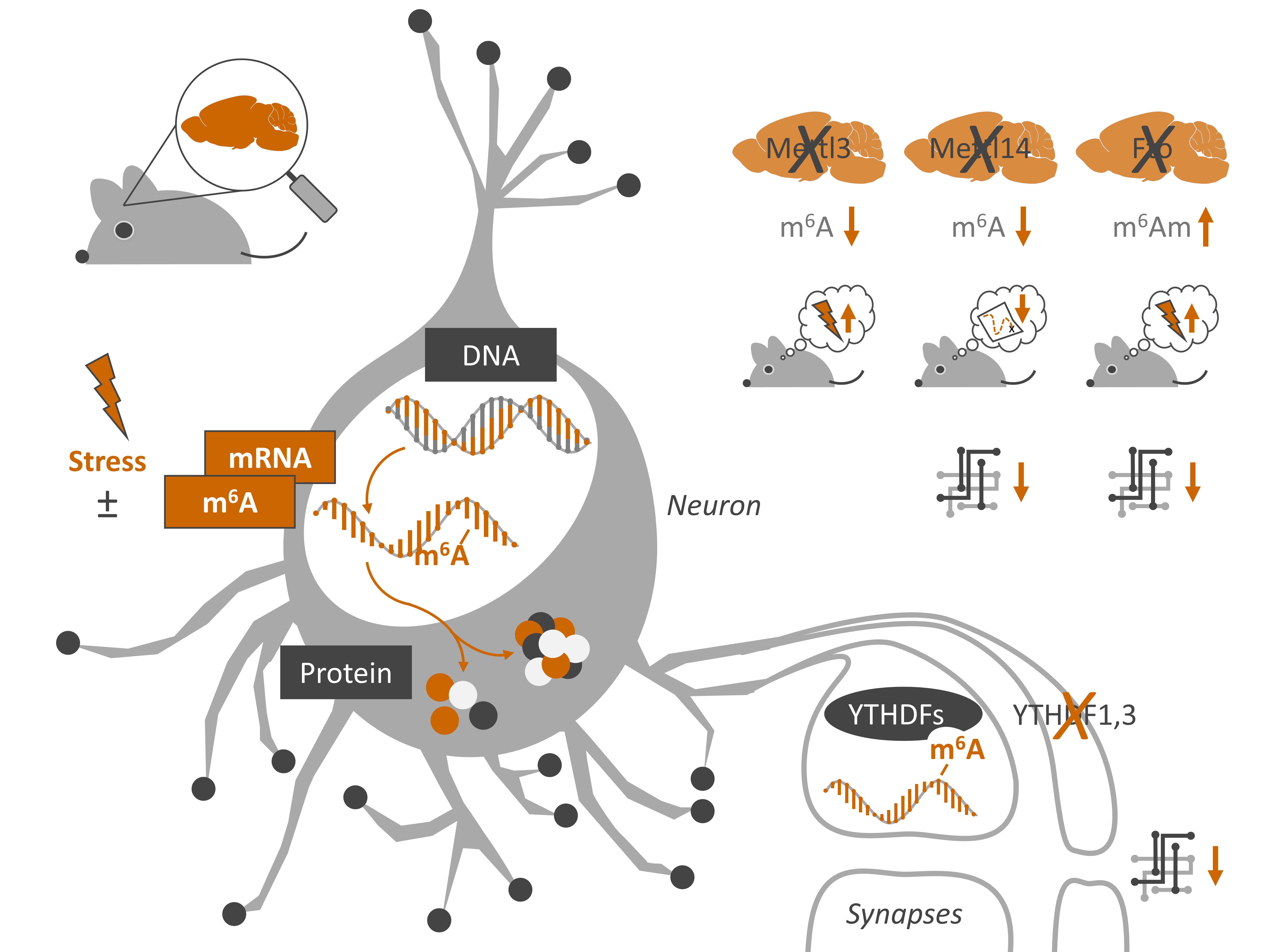
A recent study from the laboratory of Alon Chen at the Max Planck Institute of Psychiatry (Germany) examined the role of m6A in the brain in the context of stress response. They used m6A/m-seq to examine mouse cortex mRNA 4 hours after 15 minutes of acute restraint stress. They also and explored its functional role by assaying neuronal function in Mettl3 and Fto conditional knockouts. The authors were careful to point out that the antibody used cannot distinguish between m6A and the chemically similar N6,2-O-dimethyladenosine (m6Am), so they interpreted their results as a mixture. Here’s what they found:
- About half of expressed mRNAs are m6A/m modified, each with about 2 peaks/gene
- These tend to be around the 5’UTR, stop codon, and the m6A consensus motif
- m6A/m is decreased in the pre-frontal cortex and increased in the amygdala
- Injection of stress hormones (glucocorticoid corticosterone) recapitulates this effect
- The m6A methyltransferase Mettl3 is downregulated in both regions after restraint stress
- Mettl3 or Fto loss in hippocampal neurons via Cre recombinase results in:
- Genome-wide alterations in gene expression for both models
- Decreased m6A in Mettl3 deficient neurons and increased m6A in Fto deficient neurons
- Impaired fear memory in both
- Attenuated long-term potentiation in Fto but not Mettl3 deficient mice
- Global m6A/m in blood is decreased after stress in mice and humans, but not in humans with major depressive disorder
Lead author Mareen Engel shares, “m6A mRNA methylation has been described to be critical for brain development. Here we show that, although to a much more subtle degree, it is also involved in the regulation of homeostatic processes in the adult brain. Many open questions remain and future work will elucidate the details of this regulation and the mechanisms involved.”
On the challenges of m6A-seq, Engel adds: “Several protocols have been developed recently to measure m6A mRNA methylation using lower quantities of input material. However, while those readily detect a majority of the methylation sites, precisely quantifying their methylation level still presents a major challenge. Solving the quantification problem will be crucial for understanding rather modest regulation e.g. in the adult brain.”
Overall, Engel concludes “m6A mRNA methylation represents a yet hardly described mechanism to fine-tune gene expression in the brain, for example in response to stress. Integrating this mechanism with measurements of mRNA levels, ncRNAs, and epigenetic mechanisms will aid to better understand how the brain responds to external challenges and which brain alterations are associated with the etiology of psychiatric diseases.”
Visual summary of Engel et al. 2018, Neuron (image source M. Engel)
Mettl14 m6A Methyltransferase is Necessary for Normal Neuronal Function
Another recent study used a similar approach to study the effect of reducing m6A, also via conditional knockout of Mettl3. The m6A demethylase Fto has been extensively studied in disease, it has been manipulated in mouse brain leading to altered behaviour. However, Fto has numerous other functions beyond mRNA methylation, making it difficult to isolate its effect. Mettl14 encodes a key element of the m6A methyltransferase complex, which appears to be its sole function. Complete knockout of this gene is embryonic lethal in mice. But its role has been studied via conditional knockout in neural progenitor cells where its loss disrupts cortical development and leads to premature death.
A new article from the laboratory of Xiaoxi Zhuang at the University of Chicago (USA) sought to carefully examine the effects of the loss of Mettl14 in the adult brain. Specifically, they deleted Mettl14 in two neuronal subtypes in the mouse striatum. They used a Cre recombinase coupled to D1R and ADORA2A promoters to accomplish this. To address the technical challenge of low m6A IP RNA, they chose to pool three mouse samples together as one replicate.
They found:
- Mettl14 deletion in either neuronal subtype leads to decreased m6A levels, downregulation of 740 m6A-containing mRNAs, and upregulation of 498
- The magnitude of expression changes positively correlates with the magnitude of decrease in m6A for both up- and downregulated genes
- Downregulated genes are enriched for neuron-specific compartments
- Striatum-dependent learning (sensorimotor learning on the rotarod, and response-reversal learning on the water cross maze) are impaired in Mettl14-deficent mice
- Mettl14 deletion does not affect neuronal morphology, but does alter excitability and sensitivity to dopaminergic drugs
These data show that stopping m6A deposition leads to neuronal changes, specifically functional rather than morphological ones. This suggests that m6A has a specific role in protein turnover related to synaptic firing (synaptic plasticity). The final study we look in this feature explored this question in detail.
The Role of m6A in Synapses
A specific role that m6A may play in the brain is regulation of synaptic plasticity. Synaptic plasticity is the ability of the synaptic terminals of neurons to change their molecular components, structure, and transmission efficiency in response to neuronal activity; it is a core element of neuronal circuits and brain function. Altered synaptic protein synthesis in response to persistent neuron firing is a key part of synaptic plasticity. A new paper from the laboratory of Dan Ohtan Wang at Kyoto University (Japan) sought to characterize the role of m6A in synaptic function. To enrich for mRNAs present at synapses, this group used purified synaptosomes. Synaptosomes are isolated synaptic terminals from brain tissue.
Faced with the same technical challenges of m6A-seq as the above studies, the authors developed a low-input m6A-seq protocol to assess the small quantity of synaptosomic mRNA. First, sample quantification was performed using Quant-iT™ RiboGreen®, which is a highly sensitive fluorescence-based assay to measure RNA quantity. Next, standard extraction of total RNA from the synaptosomes followed by rRNA depletion and chemical fragmentation was performed. The authors then performed an optimized immunopreceipatation with an anti m6A antibody; in short, specific buffer washes and centrifugation steps were added to a standard protocol. The final piece of the low-input adaptations was co-amplification and tagging of the IPed RNA samples during cDNA synthesis using SMARTer (Switching Mechanism at 5′ End of RNA Template) cDNA synthesis. Using these modifications, they were able to generate quality libraries with 30-50 million total reads with an average length of 136 bp.
Here’s what they found:
- 4,469 m6A peaks were enriched in 2,921 genes in synaptosomes vs homogenized whole-forebrain
- The genes are enriched for neuronal ontologies such as synapse formation
- 3% of these genes do not have synapse-specific expression, suggesting local m6A deposition mechanisms or selective mRNA trafficking
- m6A reader proteins YTHDF1, YTHDF2, and YTHDF3 are enriched in hippocampal dendrites, suggesting synapses autonomously regulate m6A
- Reduced expression of either YTHDF1 or YTHDF3 via antisense short hairpin RNAs (shRNAs) in primary hippocampal neurons causes altered spine morphology, dampened excitatory synaptic transmission, and altered cell-surface protein content
These results suggest that modification of mRNAs with m6A at synapses is involved in regulating synaptic genes which affect synaptic functioning. mRNAs specifically involved in synaptic function are both enriched, and differentially m6A methylated in synapses.
Bringing it All Together at Synapses
All three studies show that m6A regulates genes relevant to brain function and that perturbation of m6A is detrimental to neuronal function. Each study suggests that m6A is particularly important at synapses, where it regulates mRNAs that control synaptic plasticity and also impacts learning and memory. Data from each study are consistent with a direct functional relationship between m6A and synaptic regulation; however, future functional studies are needed to fully characterize this. We hope we’ve made the case for making room for m6A in the world of neuroepigenetics, because it sure looks like it’s here to stay.
Check out these articles in full in:
- Neuron, July 2018 (Stress response)
- Neuron, July 2018 (Mettl14 knockout)
- Nature Neuroscience, July 2018 (Synapses)