As Ptolemy and other noteworthy cartographers will tell you, a detailed and up-to-date map is always useful. This navigational tool has guided many intrepid explorers from familiar regions towards the unknown with the promise of discovery and adventure! In the linear world of ribonucleic acid (RNA), intrepid researchers have mapped the locations of the N6-methyladenosine (m6A) modification of messenger RNA (mRNA) and revealed links to the exciting realms of heat shock responses and differentiating pluripotent stem cells.
However, mRNA methylation can also occur at cytosine residues (5mC), although we know less about the prevalence and transcriptome-wide distribution. This prompted the lab of Alexandra Lusser (Medical University of Innsbruck, Austria) to begin the mapping of 5mC in mRNA from adult mouse brain and mouse embryonic stem cells (ESCs) in the hope that a pluripotent versus differentiated cell comparison would help to decipher functions and regulatory implications.
Their unbiased global mapping effort assessed 5mC via RNA bisulfite sequencing (RNA BS-seq) of poly(A)-enriched RNA (mRNA) followed by deep sequencing using the Illumina HiSeq platform and confirmation via mass spectroscopy and methyl-RNA immunoprecipitation (meRIP).
Overall, mapping mRNA methylation in the mouse led to some interesting new findings!
- 5mC levels and diversity are higher in ESCs than in brain for total and nuclear mRNA
- Higher 5mC per transcript rates in nuclear mRNA observed compared to total mRNA
- 5mC mapped to introns and non-annotated regions suggesting a link to splicing
- Differential expression patterns did not mediate the differential methylation patterns in ESC and brain
- Points to cell/tissue type-specific mRNA methylation
- 5mC of mRNA affects transcripts related to cell metabolism and cell/tissue-specific function
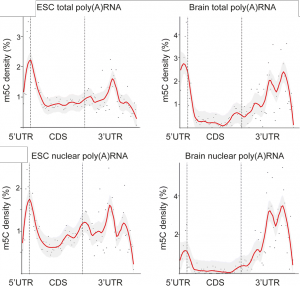
- 5mC localization on mRNA transcripts displayed striking differences
- 5mC shows cell/tissues specific mRNA localization and differences between the total and nuclear mRNA samples (See Figure)
- 5mC accumulated around the translational start codon in all samples and so may represent a common means to affect the initiation of translation
- Complex patterns in the 3’ untranslated region may permit cell/tissue-specific interactions with miRNAs or RNA-binding proteins (RBPs)
Figure adapted from Amort and Rieder et al. showing the relative levels of 5mC in various mRNA populations (CC BY 4.0)
This study represents the first survey of 5mC levels in cell types which may provide us with the ability to study the function and biological significance of this mRNA methylation mark. The next piece of the map will be to identify the enzymes responsible for targeting specific positions/regions on mRNAs for methylation with the RNA methyltransferases NSUN2 and DNMT2 the principle candidates.
So leave the familiar setting of your lab bench or office chair and go see what adventures can be had using this new 5mC mRNA map at Genome Biology, January 2017!