Every day, in labs all over the world, stem cells are changing. Sure they may lack (lineage) commitment, but these ultra-promising cellular blank canvases aren’t living sedentary epigenetic lifestyles. DNA methylation in stem cells is on the move with age and passages and you might not even know it. Sequencing still isn’t quite feasible for frequent verification of stell cells’ DNA methylation state. Some labs have used microarrays to keep tabs on things, which have been useful, but still not that feasible to implement frequently.
A new approach developed by scientists at Zymo Research uses a one-step, bisulfite-free qPCR assay to rapidly detect the DNA methylation status of key genes indicating pluripotency.
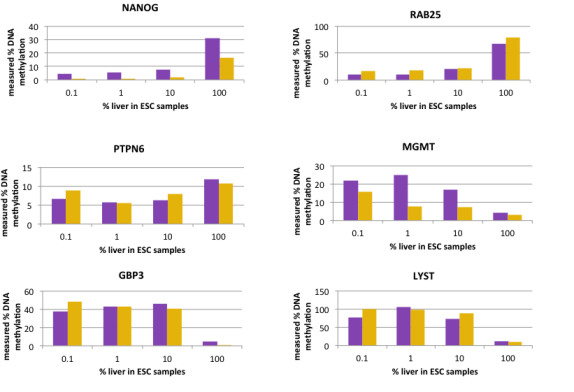
- RAB25
- NANOG
- PTPN
- MGMT
- GBP3
- LYST
Monitoring Stemness
The approach is far easier to perform than to write about (but that didn’t stop us) because there’s not much to it on the user end. You just add stem cell DNA to an appropriate well in the 96-well plate. The wells contain an optimized mix of methylation sensitive restriction enzymes, amplification reagents, buffers, and validated primers to the panel targets. After that, it’s qPCR business as usual, but here’s where it gets really interesting…
Using the unique DNA methylation states of the panel members, the researchers at Zymo used the panel to:
Detect differentiated DNA from two DNA stem cell populations (shown as purple and orange bars) using the OneStep qMethyl™ Panel
Differentiated liver cells distinguished from embryonic stem cells
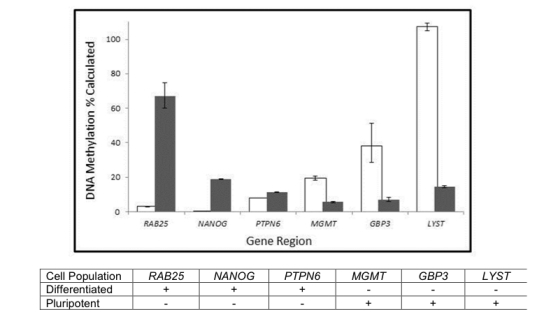
Distinguish methylation differences in a pluripotent stem cell line (shown as white bars) from a differentiated cell line (shown as grey bars
Head on over to Zymo to learn more about the OneStep qMethyl™ Stem Cell Panel.