Taking cues from ultra-talented entertainers like Justin Timberlake, methylation is out to prove it’s possible to be successful in more than just one medium. Recent research has found that methylation can pull off its’ gene expression regulatory magic at the level of RNA too. And no, we’re not talking about other RNA modifications, like 6-methyladenosine (which is highly enriched around stop codons in many mRNAs), but rather the very methylation we’ve grown to love so much over the years: 5 methylcytosine (5mC).
Interestingly, 5mC is abundant in both coding and non-coding RNAs and seems to have a role in transcript integrity, with knockouts of it’s synthesizing enzyme (Nsun2) producing intellectual disability, skin disorders, and growth retardation. Given this enormous of potential, a number of novel technologies using next-gen sequencing (NGS) have appeared quicker than a JT tune tops the charts. Here we highlight four emerging technologies as reviewed by a team from the University of Cambridge in the recent edition of Genome Biology that can help you explore RNA methylation like never before:
Bisulfite sequencing (Bs-seq):
Pros: An old fave that has been extensively used to detect 5mC in DNA has more recently been modified to work with RNA. Based on the differential chemical reactivity of 5mC compared to C, it causes the deamination of unmethylated C’s into U’s with 5mC remaining unconverted. Check out our coverage of the RNA BS-seq technique.
Cons: An important aspect of the process is the conversion rate, with a high conversion rate only being achieved by prolonged incubation under harsh chemical conditions, which cause RNA degradation. Also, as a consequence, very deep sequencing is required for low abundance transcripts.
Now let’s go into some of the emerging options that take advantage of RNA immunoprecipitation. Also be sure to check out our free RIP guide and RIP/iClIP protocols.
m5C-RNA Immunoprecipitation (m5c-RIP):
Pros: Relies soley on biology, so no harsh chemical conversion is needed.
Cons: Has not been performed in Mammalian Cells yet.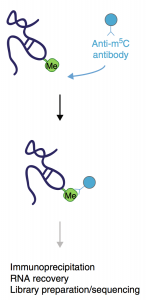
5-Azacytidine-mediated RNA Immunoprecipitation (Aza-IP):
A clever exploitation of the formation of a covalent bond between a modified RNA base and the RNA methylase (NSun2).
Pros: It lets you catch RNA and it’s significant enzyme in the act.
Cons: It is unclear why and how this conversion occurs. It also relies on the chemical incorporation of 5-aza into the entirety of the transcriptome prior to isolation of protein-RNA complexes, which “may compromise RNA stability and integrity”, while the analogue also incorporates itself into DNA
Methylation-individual Nucleotide Resolution Crosslinking Immmunoprecipitation (miCLIP):
A cunning use of a point mutation that leaves the methylating enzyme (NSun2) incapable of leaving its’ former RNA ‘associate’ after the act.
Pros: A more ‘natural’ way to catch RNA methylation in action.
Cons: It appears that less than 10 % of the methylome is directly dependent on NSun2
See more detailed figures and descriptions of RNA methylation analysis methods at Genome Biology, December 2013

