Guess what? It seems that blue light has a lot more to offer than just helping with your winter time blues. It could also be just what your transcriptional activation system needs. Synthetic biology has a lot to offer omics beyond genome editing and recent work from multiple groups is putting Cas9 in a different spotlight.
CRISPR/Cas9 can be modified to target RNA for editing and avoid corresponding DNA, but it can also be used to take control of gene expression. A number of modified genome editing systems have arisen to this challenge, with modifications that can take advantage of the targeting system paired to an inactivated nuclease, which is fused to an effector of interest. This allows for targeted epigenomic modifications or transcriptional control via activation or even just repression by some old school steric hindrance.
Light-inducible Transcriptional Effectors
However, just like genome editing, editor-based transcriptional activation systems have got some obstacles to overcome. Two of the most challenging are the delivery systems and induction. Previously, Feng Zhang and his lab created a system termed LITE (light-inducible transcriptional effectors), which is based on the earlier transcription activator-like effector nuclease (TALEN) genome editing technology to tackle the issue of induction.
This neat little system comes at you right from Arabidopsis, with a duo of protiens:
- Light-sensitive cryptochrome 2 (CRY2).
- Binding partner CIB1.
- The binding, which leads to their heterodimerization, is rapid and reversible, you’ve just gotta shine a little blue light.
To create the LITE system, Silvana Konermann along with others researchers from the Zhang lab fused the TALE DNA binding domain to CRY2 (thus giving sequence specificity by protein binding to DNA) and the VP64 activator was fused to CIB1. Then when CRY2 gets a little over excited it binds to CIB1 and drives expression.
Light-Activated CRISPR-Cas9 Effectors
This system has now been tweaked to use CRISPR/Cas9 as shown in two recent papers. By using a combination of optogenetics and CRISPR/Cas9, the teams were able to have their systems on call for some spatio-temporal control.
In the CRISPR/Cas9 version of the optogenetic system:
- CRY2 is fused to the transactivation domain (either p65 or VP64).
- C1B1 is fused to dCas9, the deactivated Cas9 nuclease from CRISPR.
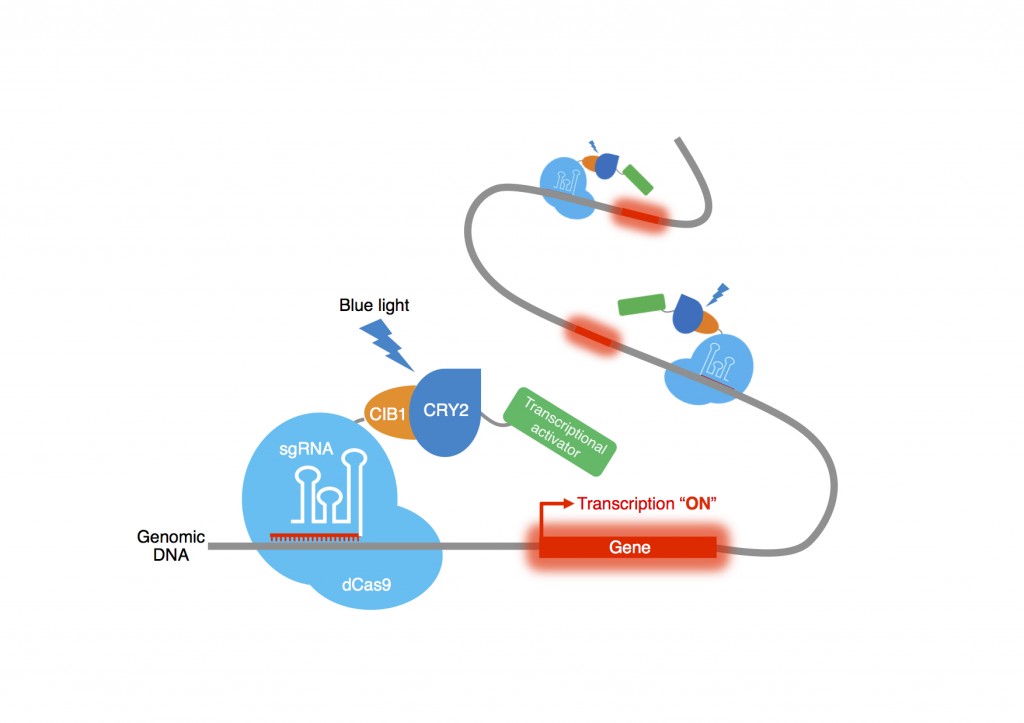
A CRISPR/dCas9 based Photoactivatable Transcription System. Image courtesy of Dr. Moritoshi Sato http://satolab.c.u-tokyo.ac.jp
The CRISPR version goes by the flashy name of LACE (light-activated CRISPR-Cas9 effector) or LITE 2.0, depending on the pub. It also has a few more improvements than just swapping out the TALE for dCas9 and was shown off in multiple chosen endogenous mammalian genes. The two systems, developed independently by researchers in the labs of Moritoshi Sato and Charles Gersbach, only appear to differ by the CRY2 domains per a dCas9 and also the activator of choice.
Ultimately, optogenetics provides the perfect ‘on’ switch for an on-the-fly fusion of dCas9 and a transactivation domain that gets the expression going. By using the dCas9 fusion the teams were able to take advantage of the excellent sgRNA targetting system that has an almost infinite number targets and is much cheaper than engineering targeting proteins, all the while not chopping away at the gene of interest. We’re left wondering how much longer until one of our favorite epigenetic modifiers joins the light show.
Go and see what the blue light does to your favourite genes in Chemical Biology, February 2015 and Nature Chemical Biology, February 2015.



