Hi-C-based three-dimensional chromatin conformation analysis is currently living the high-life, with multiple high-impact studies recently employing this high-flying molecular technique to gain new insight into the cell cycle, fertilization, and reprogramming.
Now, researchers under the high command of Boyan Bonev and Giacomo Cavalli (Université de Montpellier, France) have applied this high-class technique to understand how the spatial proximity of functional chromatin elements relates to gene expression and cell fate via the creation of high-resolution Hi-C maps of neural cell differentiation.
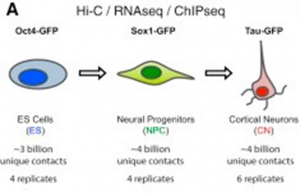
And when we say high-resolution, we mean high-resolution! The authors sequenced over 40 billion paired-end reads, resulting in around 17 billion uniquely mapped contacts derived from sorted mouse embryonic stem cells (ESCs) and ESC-derived neural progenitor cells (NPCs) and cortical neurons to act as a model of sequential neuronal differentiation in vitro (See Figure).
This high-fidelity dataset facilitated the creation of high-end 3D chromatin conformation maps at a maximum resolution of 750 bp, the highest of any reported study. The authors then correlated these findings with chromatin modification analysis (ChIP-Seq and reappraisals of ENCODE datasets) and RNA sequencing (RNA-Seq) to explore the relationship between gene expression, the epigenome, and 3D genome conformation.
So what did this high-resolution Hi-C analysis tell these highly motivated researchers?
- In vitro modelled neural development led to the dynamic global reorganization of chromatin interactions
- Topologically associating domains (TADs) reduced in number but increased in size
- The number of long-range active domain (A) interactions decreased, while the number of inactive domain (B) interactions increased
- Strong polycomb-mediated interactions present in ESCs destabilized
- Gene transcription highly correlated to chromatin domain insulation and long-range interactions
- All cell types exhibited long-range contacts between different active gene promoters and also between the bodies of different exon-rich active genes
- TAD boundary formation at or close to active gene promoters also increased during differentiation suggesting that novel borders can form at the promoters of developmentally regulated genes
- In vivo analysis of neural development via the purification of NPCs and cortical neurons from the mouse neocortex (∼3 billion uniquely aligned contacts per cell type) also demonstrated global chromatin reorganization
- Neural transcription factors mediate dynamic and cell-type specific enhancer-promoter interactions that are constrained by TADs
- These interactions are established concomitantly with gene expression and become disrupted following gene repression
Overall, this remarkably high-resolution high-impact study represents a treasure trove of data concerning how three-dimension chromatin conformation changes during neural differentiation and how this correlates to gene expression and in vivo function.
To gain a high-resolution view of this new Hi-C study, get off your high horse (!) and head over to Molecular Cell, October 2017.
Schematic representation of the in vitro neural differentiation system (CC BY 4.0)